Barr Lab
Our research: Negative stranded RNA viruses
We are interested in a broad range of research topics concerning several negative stranded RNA viruses. These topics range from investigating the structural biology of individual viral proteins, interactions between viral and host cell components, through to how virus infection affects cellular processess and pathways on a whole-cell basis. The viruses we currently study include several bunyaviruses (Crimean-Congo hemorrhagic fever virus, Hazara virus, Bunyamwera virus, Schmallenberg virus and others), some arenaviruses (LCMV, Pichinde and Lujo viruses) and the non-segmented human respiratory syncytial virus.
The links on the left will direct you to individual pages describing each of these viruses along with some current research projects, and the people doing the work.
We often have PhD studentship opportunities for talented and enthusiastic scientists. These opportunities come and go throughout the year, and more details can be found at findaphd.com, on the FBS web pages, or on the 'PhD opportunities' page that can be accessed using the menu on the left. Please contact me directly if you are unsure how to proceed or need more information. Below are some examples of the type of work we do:
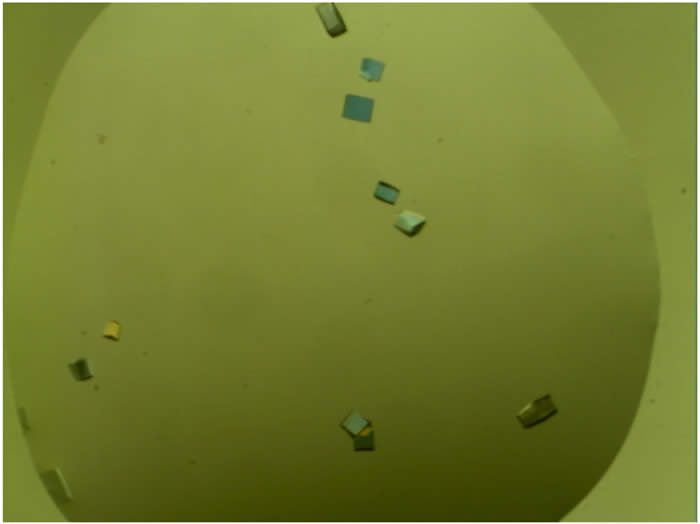
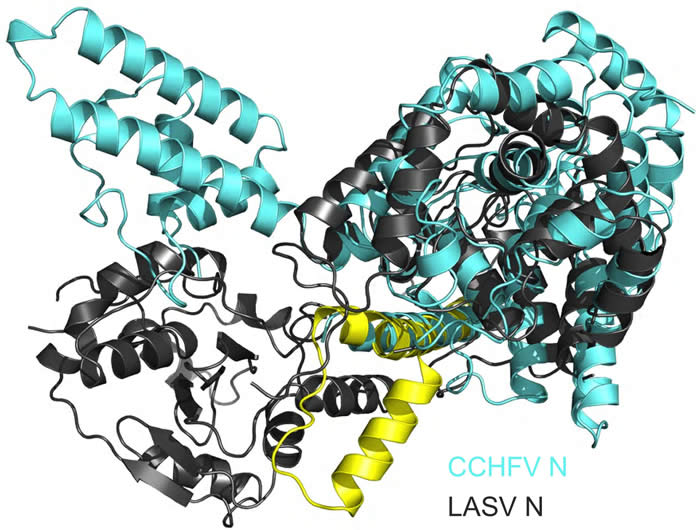
1). Above is an example of data generated during a recent PhD research project (from left-to-right): Crystals of the CCHFV N protein used to solve its high resolution structure. Alignment unexpectedly revealed the CCHFV N protein was more closely related to the corresponding protein from an arenavirus (Lassa virus) rather than any other bunyavirus. These two structures are shown in overlay. This was supported by phylogenetic analysis of both N and L proteins of segmented negative stranded RNA viruses, revealing unexpected evolutionary relationships between them.
 .
.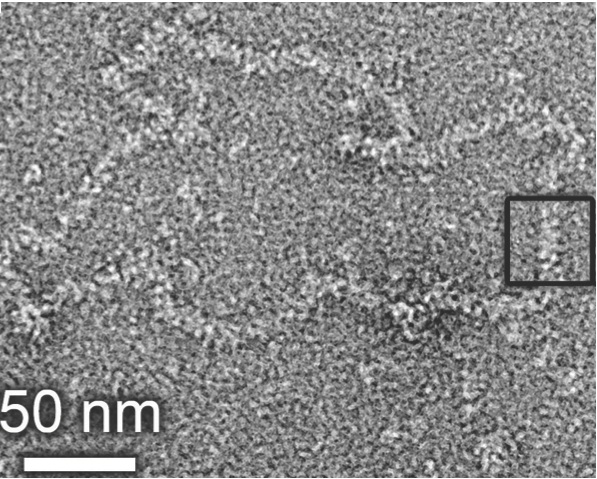
2). Above is a representation of the crystal structure of the Bunyamwera virus N protein in complex with RNA. The N protein forms a tetramer in both solution and in crystals, and is shown here with electrostatic surface rendering. The genomic RNA is sequenstered deep inside a positively charged (blue) cleft, where it is protected from environmental insult, and sentinels of cellular innate immunity. However, this raises very interesting questions about how the RNA is accessed by the polymerase during RNA synthesis. The image to the right shows a negative stained EM of a purified Bunyamwera RNP. The width of the RNP within the boxed region is consistent with the diameter of the tetramer, suggesting that this RNP conformation can be built from stacked protohelical tetramers.
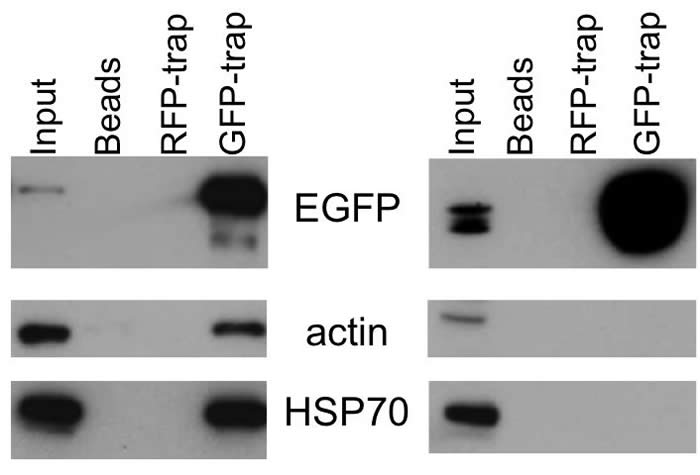
3). Shown above are data that identify host cell proteins that interact with the CCHFV and HAZV nucleocapsid proteins during their respective life cycles. These interactions were first detected by using quantitative proteomics of immunoprecipitated N protein, extracted from cell lysates. The most abundant group of cell proteins identified were cellular chaperones of the HSP70 and HSP90 families, as well as their DnaJ co-chaperones, and some cytoskeletal components. The N-HSP70 and N-actin interaction were confirmed by western blotting (top image), and immunofluorescence (bottom image) was used to confirm a high degree of colocalization in both transiently transfected and virus infected cells. Blocking the HPS70-N interaction using HSP70 inhibitors reduced virus yields by up to 1000 fold.
