Research facilities and Technical services

- Faculty of Biological Sciences
- Facilities
Facilities
The Faculty of Biological Sciences has a number of research facilities which are open to all researchers at the University of Leeds, other academic institutions, and industry. Their purpose is to support world-class research at the interface of chemistry, biology, physics, medicine and engineering. As well as state-of-the-art instrumentation, each facility is staffed by experts in their field who can advise on all aspects of application of the techniques to your research including experimental design, training, data analysis and experimental assistance.
Our ten facilities cover the broad research themes of biophysical characterisation, structural elucidation and cellular visualisation. Together they provide a full pipeline for preparation and complete characterisation of systems from single molecules, to macro-molecular complexes, to cells. For more information, use the links on this page to explore the research facilities within each theme. All University of Leeds users can find out more about accessing our Research Facilities via our SharePoint site at https://leeds365.sharepoint.com/sites/FBSResearchFacilities/
£17 million
Astbury Biostructure Investment
£630k
Protein Production Facility investment
£2.5 million
Investment in Mass Spectrometry equipment
Biophysical techniques
Wide-ranging techniques to answer an array of technical questions, such as discovering interactions between molecules, investigating conformational changes, and determining binding kinetics.
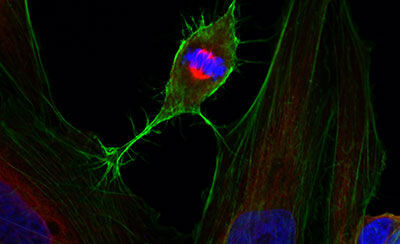
Cellular techniques
Diverse equipment for producing high quality recombinant proteins and imaging cells and tissues, including wide-field, confocal or super-resolution fluorescence systems, electron microscopy, flow cytometric analysis, and support with sample preparation.
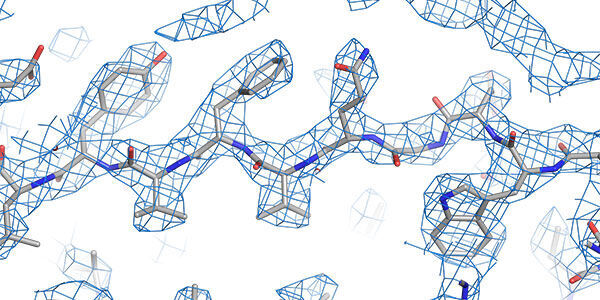
Structural techniques
World leading state-of-the-art facilities for investigating the structure and conformation of biomolecules down to the atomic scale.

Technical Services
Our world-leading research facilities are supported by our dedicated Technical Services team, who work across all areas of our research.

